Frequently Asked Questions

|
|
1. ProTox 3.0
1.1 ProTox 3.0
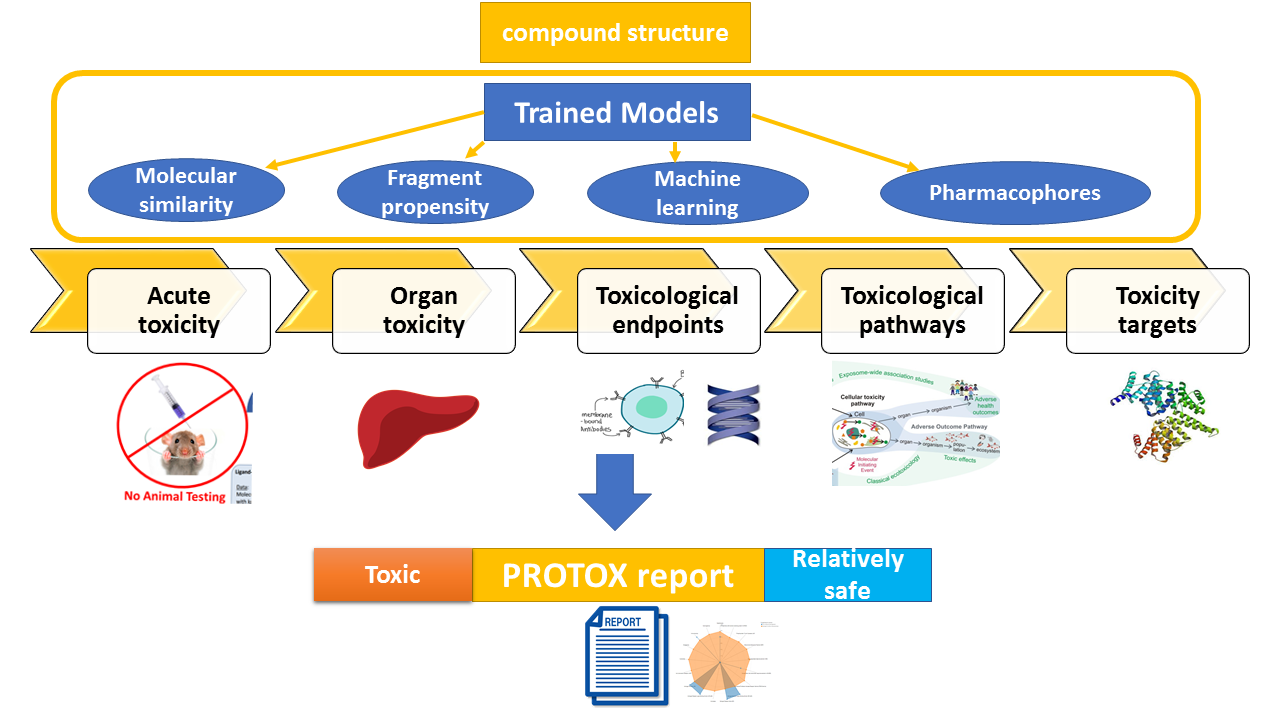
ProTox 3.0 is a virtual toxicity lab enabled to academic and non-commercial users via a web server, for the prediction of multiple toxicological endpoints related with a chemical structure. ProTox 3.0 contains computer-based models trained on real data ( in vitro or in vivo ) to predict the toxic potential of the existing and virtual compounds. The acute toxicity class as well as different endpoints are calculated for an input compound based on chemical similarities to toxic compounds and trained machine learning models. ProTox 3.0 envisage itself as a freely-available complete computational platform for in silico toxicity prediction for toxicologist, regulatory agencies, computational chemist and medicinal chemist.1.2 Purpose of ProTox 3.0
ProTox 3.0 is a free web service and the user can create a toxicity prediction for an input compound within a few minutes. Computational toxicity predictions can help to reduce the amount of animal experiments and save animal lives. ProTox 3.0 incorporates molecular similarity and machine-learning models for various toxicity endpoints. A novelty of the ProTox webserver is that the prediction scheme is classified into different levels of toxicity such as oral toxicity (acute rodent toxicity), organ toxicity (hepatotoxicity), toxicological endpoints (such as mutagenicity, carcinotoxicity, cytotoxicity and immunotoxicity (B cell growth inhibition)), molecular initiating events (MOE) toxicological pathways (AOPs) and toxicity targets (Novartis off-targets) thereby providing insights into the possible molecular mechanism behind such toxic response.Predict compound toxicity
2. ProTox-Server
2.1 System Requirements
We recommend a recent version of Mozilla Firefox or Google Chrome, though the site should also be usable with Microsoft Internet Explorer (10,Edge) or Apple Safari. JavaScript has to be enabled to use all the features of the site. Depending on your browser and security settings, certain features like the Radar Chart might request you to permit the use of local browser storage for session data.2.2 Server Implementation
ProTox 3.0 data is stored in a relational MySQL database. To handle the chemical information within the database, the MyChem package is used. For most of its functions, MyChem relies on the Open Babel toolbox. The website back-end is built using PHP; web access is enabled via the Apache HTTP Server. As an agile key/value store, Redis, is employed for queueing and assessing API requests.2.3 Using the API
For advanced users, data can be queried using a simple POST interface with a suitable language of your choice. Below, a short introduction and sample code in Python (Version 3.6 or newer) is provided. Please note that for single queries, the script is slower than the website, as it is set to allow several users a chance to queue their requests. The more models you require, the longer the query intervals take due to computation time. A source IP is allowed a maximum of 250 API queries a day.You can download this script to your local computer and use it, or write your own with the script as a reference : Sample API Script
To run the script, you would need to install python (3.6 or newer) on your system, and invoke your command line (either via cmd on windows, or opening a terminal on linux or mac os). The interface allows you to query by name (fulfilled via PubChem search) or canonical SMILES string. As a minimum, you need only enter one or more such identifiers (separated by comma).
If you prefer no status outputs save errors, use the -q command line switch.
python3 protox3_api.py aspirin,vorinostat
Additional data can be supplied using command line switches, from specifying the input type (if you want to input canonical SMILES), to selecting the models (you can see a full list of models either in the Toxicity Model Report Table, in the Shorthand column, or in the script itself, in the ALL_MODELS declaration)
python3 protox3_api.py -t smiles -m "acute_tox tox_targets ALL_MODELS" -o out.csv "CCC(=C(C1=CC=CC=C1)C2=CC=C(C=C2)OCCN(C)C)C3=CC=CC=C3"
PLEASE NOTE : As seen below, add quotation marks if you include a SMILES string. Likewise,use quotes around the whole query if split drugnames (two words) occur.
The API by returns data in the form of a CSV file. Therefore the data is human readable but can also be easily processed in most languages and many software. The following columns and information is provided in each response written to your outfile:
input : If using name input type, the compound name, if using canonical SMILE input type, the input smiles
type : Either "acute toxicity", "toxicity model" or "toxicity target"
acute_tox : If selected, the acute toxicity prediction with LD50, toxicity class and prediction accuracy data
tox_models : If one or more models selected data with name, prediction and prediction confidence
tox_targets : If selected,toxicity targets with pharmacophore fit values
Target : The predicted type, model or target of the corresponding prediction
acute_tox : Either LD50 or tox_class
tox_models : Shorthand name of the model
tox_targets : Name of toxicity target
Prediction : The predicted outcome
acute_tox :
LD50: predicted ld50 in mg/kg
tox_class : predicted toxicity class (1-6)
tox_models : Boolean value if activity or inactivity is predicted (1=Active, 0=Inactive)
tox_targets : Probability class for binding from 0 to 3 (0=no binding, 3=probable binding), color-coded on website
Probability : criterion differs depending on the selected target
acute_tox : average similarity in % (float 0-100)
tox_models : float value from 0 to 1 giving confidence of the above result
tox_targets : float value from 0 to 1 giving the average pharmacophore fit score
Please note the output is intended to be machine-readable. To inspect it manually, using a JSON-Viewer like Stack.hu JSON viewer or Code Beautify JSON Viewer is recommended.
However, otherwise, the website itself is more suitable to such visualization.
3. Biological Background
3.1 Toxicity prediction
The investigation of the absorption, distribution, metabolism, excretion and toxicity, the so-called ADMET properties of a compound, is a crucial step in the drug development process. Before a drug candidate proceeds into clinical trials, its ADMET properties have to be determined. Usually, toxicities are investigated in animal experiments which are time-consuming and take animal lives. In silico toxicity predictions are a fast and inexpensive alternative to animal experiments. They rely on known toxicity data which is used to develop a model capable of predicting toxicities of new compounds. On the other hand, mechanism-based prediction and evaluation of chemical toxicity is still an evolving science, and such understanding is important for the development of drugs as well as regulatory decisions. A particular compound can be active for multiple toxicity endpoints. A chemical that interacts with a protein as an off-target, can also interact with multiple proteins with different affinities, consequently it can activate different signalling pathways or interact with multiple functional pathways. The signaling or functional pathways that are perturbed may have overlapping connectivity, resulting in synergistic or canceling system consequences. Similarly this can extend to across organ, tissues, cellular levels of connectivity, resulting in servere and strong toxic profiles (P.F. Bai et al. 2013).3.2 Toxicity classes
Based on the severity of their effect, compounds can be classified into different toxicity groups (classes). As explained here, our webserver uses the GHS toxicity classification whereby compounds are divided into 6 classes - 5 classes representing different grades of toxicities as well as the non-toxic class.3.3 Toxic fragments
Toxic fragments were generated using ROTBOND and RECAP method. There were about 37000 distinct fragments created from toxic and non-toxic compounds present in the training data set. Based on statistical analysis on the occurence of each fragment, fragments were further classified as specific fragments with respect to the toxicity classes. Additionally, these specific fragments are used to predict the toxic class for the input molecule. Some examples of molecules containing the specific toxic fragments are Perfluoroterephthalonitrile, Chloroflurazole, Benzimidazole, 4,5,7-trichloro-6-nitro-2-(trifluoromethyl) etc. Few examples of specific toxic fragments present in our training dataset are shown below: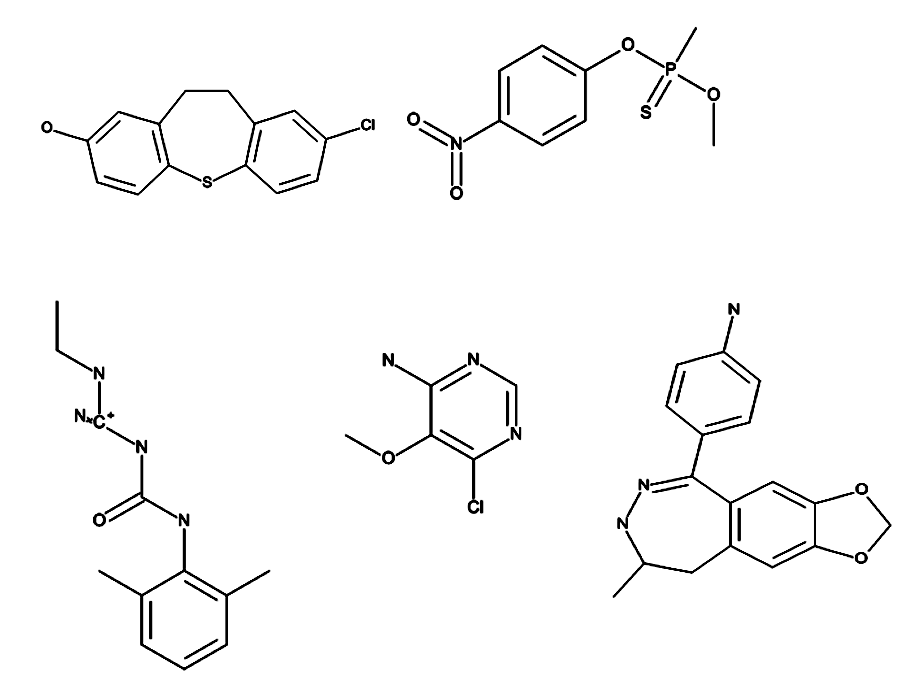
3.4 Acute Toxicity
Acute toxicity describes the adverse effects of a substance that result either from a single exposure or from multiple exposures in a short period of time (e.g. less than 24 hours). The acute oral toxicity prediction results are based on the analysis of 2D similarities and the recognition of toxic fragments in approximately 38, 000 unique compounds with known oral LD50 values measured in rodents.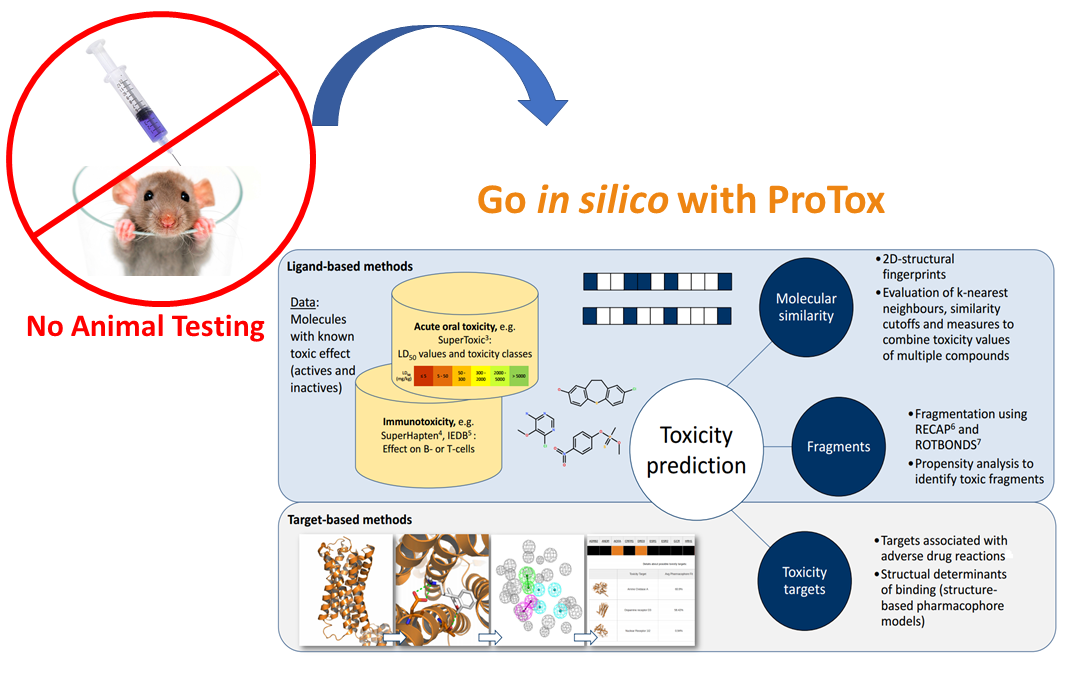
3.5 Organ Toxicity
Chemicals that can cause adverse effects or disease states manifested in specific organs of the body is defined as organ toxicity.Hepatotoxicity refers to liver dysfunction or liver damage that is associated with an overload of drugs or xenobiotics. The liver cell injury can be due to a multitude of causes including drugs, toxins, herbal and dietary supplements, and other agents.
The organ toxicity prediction results are based on the trained machine learning model using Random Forest Classifier and discriminative features. Additionally, future considerations of additional organ based toxicities such as cardiotoxicity, neurotoxicity are planned.
3.6 Toxicological Endpoints
ProTox 3.0 currently includes methods for prediction of four toxicological endpoints such as cytotoxicity, mutagenicity, carcinogenicity and immunotoxicity.Chemicals that change the genetic material, usually DNA of an organism are defined as mutagen and the adverse effects is called mutagenicity.
Chemicals that can cause cells to become cancerous by altering their genetic structure so that they multiply continuously and become malignant are called carcinogens and the adverse effects is called carcinogenicity.
Chemicals that alters the functioning of the immune system upon exposure are called immunotoxins and the adverse effect is called immunotoxicity. The current immunotoxicity model is based on B cell growth inhibition. Additonal model on T cell inhibition will be added soon.
All the models are based on machine learning methods and the results are predicted with a confidence score.
3.7 Toxicological Pathways
Toxicology in the 21st Century (Tox21) is a federal collaboration among EPA, NIH, including National Center for Advancing Translational Sciences and the National Toxicology Program at the National Institute of Environmental Health Sciences, and the Food and Drug Administration.According to the Tox21 Consortium, chemical compounds might have the potential to disrupt processes in the human body that may lead to negative health effects.
The researchers at Tox21 consortium have tested 10,000 environmental chemicals (called the Tox21 10K library) for their potential to disrupt biological pathways that may result in toxicity, this associated pathways are called adverse outcome pathways (toxicological pathways). The Tox21 data challenge which was hosted in the year 2014, consisted of 12 pathways based on cellular assays, under two types of pathways. The idea behind the approach is that a chemical compound when interacts with the receptors, enzymes etc (either activate/inhibit) can result in perturbation in the biological pathways and thereby disrupt the cellular process causing cell death.
The two pathways namely defined as
1)Nuclear Receptor Signalling Pathways (7 pathway assays)
2)Stress Response Pathways (5 pathway assays)
Important: Many compounds in Tox21 10K library have shown cytotoxicity in a lower concentration than the concentration needed to interact with a receptor. As mentioned in the paper (Judson et al. 2013), some of the compounds might kill the cells before having any action on the receptor. Since these information are not considered in the model training (due to data/information inavailability), the users are requested to keep this information in mind and output of these models should be considered with caution.
More information on Tox21 pathways can be found here.
3.8 Molecular Initiating Events
Compound toxicity can be caused by many different mechanisms. Adverse toxicological effects are often categorized as chemical-based, on-target, or off-target effects. Chemical-based toxicity is defined as toxicity that is related to the physicochemical characteristics and structure of a compound and its toxic effects on cellular organelles, membranes, and/or metabolic pathways. On-target refers to exaggerated and adverse pharmacological effects at the target of interest in the system (Rudmann et al. 2013). It is imperative to use the toxicological and biological data on the target to form testable hypotheses related to whether a toxicity is chemical-based, on-target, or off-target. To understand the underlying mechanism, it is important to consider the macromolecular targets to which a compound binds. Some targets are important for the therapeutic effect of the drug compound. Other targets, so-called 'off-targets' or 'tox targets' are responsible for adverse drug reactions and toxicities associated with a drug compound.Which toxicity targets are we considering?
A list of all toxicity targets is available here.
Currently, only 14 MIE targets for which chmeical structures of ligand are avaialble, are considered for prediction. These include:
- Glutamate N-methyl-D-aspartate receptor (NMDAR),
- alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionate (AMPAR),
- kainate (KAR),
- Na+/I− symporter (NIS,
- Acetylcholinesterase (AChE),
- Ryanodine-sensitive Ca2+ channels (RyR),
- Thyroid hormone receptor alpha (THRα),
-Thyroid hormone receptor beta (THRβ),
- Thyroperoxidase (TPO),
- Transtyretrin serum binding protein (TTR),
- onotropic GABA receptors (GABAR),
- the pregnane X receptor (PXR),
- The constitutive androstane receptor (CAR),
- NADH-quinone oxidoreductase (NADHOX),
- Voltage-gated sodium channels (VGSC).
However, future considerations of additional targets as well as other types of pharmacophore models are planned.
3.9 Toxicity targets
Adverse Outcome Pathways (AOPs) provide information on relevant molecular initiating events (MIEs) and key events (KEs) that could inform the development of computational alternatives for these complex effects. (Gadaleta et al. 2022). The MIEs of existing AOP networks of developmental and adult/ageing neurotoxicity were modelled to predict additional neurotoxic potentials of the chemical compounds. It is imperative to use the toxicological and biological data on the target to form testable hypotheses related to whether a toxicity is chemical-based, on-target, or off-target. To understand the underlying mechanism, it is important to consider the macromolecular targets to which a compound binds. Some targets are important for the therapeutic effect of the drug compound. Other targets, so-called 'off-targets' or 'tox targets' are responsible for adverse drug reactions and toxicities associated with a drug compound.Which MIE targets are we considering?
Molecular Initiating Events associated with Developmental Neurotoxicity, adapted from Spînu et al. [45] and Li et al. [27] Tox targets have been defined according to the Novartis in vitro safety panel of targets associated with adverse drug reactions (Lounkine et al. 2012). 73 toxicity diverse toxicity targets are considered, including transmembrane proteins as well as intracellular receptors. A list of all toxicity targets is available here.
Currently, only protein targets for which experimental structures of human protein-ligand complexes have been solved, are considered for prediction. These include:
- Adenosine A2A receptor (2 toxicophores),
- Adrenergic beta 2 receptor (7 toxicophores),
- Androgen receptor (46 toxicophores),
- Amine oxidase (3 toxicophores),
- Dopamine D3 receptor (1 toxicophore),
- Estrogen receptor 1 and 2 (195 and 32 toxicophores),
- Glucocorticoid receptor (7 toxicophores),
- Histamine H1 receptor (1 toxicophore),
- Nuclear receptor subfamily 1 group I member 2 (97 toxicophores),
- Opioid receptor kappa (4 toxicophores),
- Opioid receptor mu (1 toxicophore based on homology model),
- cAMP-specific 3',5'-cyclic phosphodiesterase 4D (152 toxicophores),
- Prostaglandin G/H synthase 1 (1 toxicophore),
- Progesterone receptor (9 toxicophores).
However, future considerations of additional targets as well as other types of pharmacophore models are planned.
How are toxicity targets predicted?
Tox targets are represented in form of pharmacophore models. For each toxicity target, a set of of pharmacophore models is generated and validated using a set of active compounds and property-matched decoys. Only pharmacophores receiving a good validation result are used further. The fit value of a pharmacophore to a compound is then used as an indicator of the strength of binding to a specific target.
3.10 Metabolism
Compound toxicity can be caused by many different mechanisms. Drug metabolism via the cytochrome P450 system has emerged as an important determinant in the occurrence of several drug interactions that can result in drug toxicities, reduced pharmacological effect, and adverse drug reactions. Recognizing whether the drugs involved act as enzyme substrates, inducers, or inhibitors can prevent clinically significant interactions from occurring. ProTox 3.0 platforms predicts six major CYPs isoforms, including 1A2, 2C9, 2C19, 2D6, 2E1 and 3A4, that are responsible for more than 90% of the metabolism of clinical drugs. For more detailed predictions and literature curated details on known cytochrome interaction network of approved drugs please check SuperCypPred here.CytochromeP450 (CYPs) enzymes mediated drug metabolism influences drug pharmacokinetics and results in adverse outcomes in patients through drug-drug interaction (DDIs). To understand the underlying mechanism, it is important to consider the macromolecular targets to which a compound binds. Some targets are important for the therapeutic effect of the drug compound. Other targets, so-called 'off-targets' or 'tox targets' are responsible for adverse drug reactions and toxicities associated with a drug compound.
Which CYPs enzymes are we considering?
Currently, only sic CYPs molecular targets are considered for prediction. These include:
- CYP1A2: CYP1A2 is expressed in the liver and accounts for approximately 13% - 15% of the total CYP content, contributing to the metabolism of approximately 4% of marketed drugs. CYP1A2 preferentially oxidizes aromatic hydrocarbons as well as heterocyclic and aromatic amines and plays an important role in the metabolism of several clinical drugs, including analgesic, antipyretic, antipsychotic, antidepressant, anti-inflammatory, and cardiovascular drugs. CYP1A2 has been reported to catalyze N-hydroxylationof pre-carcinogenic heterocyclic amines to carcinogenic compounds. Therefore, in addition to predicting DDIs, it is important to understand CYP1A2 inhibition while researching on carcinogenesis.
- CYP2C9: The CYP2C9 family accounts for approximately 20% of hepatic P450s, and CYP2C9 is responsible for the hepatic clearance of 15% of clinically relevant drugs (including phenytoin, tolbutamide, and warfarin) as the first step in drug clearance, limiting drug oral bioavailability. CYP2C9 inhibitors include fluvastatin, flu- voxamine, zafirlukast, and antifungal imidazole compounds (mi- conazole, fluconazole, and sulconazole).
- CYP2C19: CYP2C19 is an essential member of the CYP450 superfamily and it contributes about 16% of total hepatic content. CYP2C19 is the principal enzyme involved in the hepatic metabolism of drugs such as antimalarial (proguanil), oral anticoagulants (R-warfarin), chemotherapeutic agents (cyclophosphamide), anti-epileptics (S-mephenytoin, diazepam, phenobarbitone), antiplatelets (clopidogrel), proton pump inhibitors (omeprazole, pantoprazole, lansoprazole, rabeprazole), antivirals (nelfinavir), and antidepressants (amitriptyline, clomipramine).
- CYP2D6: CYP2D6 metabolizes approximately 30% of all marketed drugs, including antiarrhythmics, antidepressants, antipsychotics, beta- blockers, and analgesics; although it accounts for only 2%-4% of all human hepatic CYPs. CYP2D6 is a polymorphic P450 iso- form, in which the active enzyme is absent in 5%-10% of Caucasians and 1% of Asians. Therefore, much emphasis is placed on CYP2D6 metabolism and its potential for clinically relevant drug interactions early in the drug discovery process.
- CYP3A4: CYP3A4 is the most abundant human hepatic CYP isoform and is responsible for the metabolism of approximately 50% of known drugs, including cyclosporine, testosterone, dextromethorphan, diazepam, and midazolam. The inhibition of CYP3A4 by co- administered drugs is shown to result in clinically adverse DDIs owing to the decreased systemic clearance of CYP3A4 substrates and rapid and unexpected increases in plasma concentrations. Indeed, most DDIs that result in the withdrawal of drugs that are already available in the market are caused by CYP3A4 inhibition. Therefore, the early identification of potential CYP3A4 inhibitors is required to minimize the risk of clinically relevant interactions.
- CYP2E1:CYP2E1, or cytochrome P450 2E1, is an enzyme involved in the metabolism of various substances in the body, particularly xenobiotics (foreign compounds) such as drugs and environmental toxins. It is a member of the cytochrome P450 superfamily of enzymes, which play a crucial role in the detoxification and elimination of a wide range of substances. CYP2E1 is primarily found in the liver, but it can also be present in other tissues. CYP2E1 is involved in the activation of certain toxic substances. For example, it can convert some chemicals into reactive intermediates that may contribute to liver toxicity. This enzyme can also be involved in drug interactions, as its activity can affect the metabolism of drugs that are substrates for CYP2E1.
However, future considerations of additional CYPs taergets as well as other types of metabolism enymes predictions are planned.
3.11 Performance analysis
The performance of a binary prediction method can be assessed retrospectively, using a validation set with known activities. For example, when considering the prediction whether a compound is in toxicity class 1 or is not in toxicity class 1, the following four values can be determined: the number of true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN). From these numbers, the following performance parameters can be calculated:1) Sensitivity/ Hit rate/ Recall/ True positive rate: TP/all positives in validation set
2) Specificity/ True negative rate: TN/all negatives in validation set
3) Precision/ Positive predictive rate: TP/(TP+FP)
4) Balanced accuracy: (Sensitivity+specificity)/2
5) Kappa index measures the quality of the binary classification models
6) The area under the curve (AUC) of a receiver operating characteristic (ROC): curve plots the sensitivity versus 1-specificity at different threshold
7) Identification of frequent features in active and inactive compounds: To analyse the important and frequent features in active and inactive compounds. The percentage of occurrences of each feature from Morgan fingerprint (2,048 bits) in active and inactive compounds was calculated. The relative frequency of important features for a class (e.g., active) were calculated taking not only the feature position and occurrence within the active class into account but also the relative feature frequency of that feature in the inactive class and vice versa. The average relative frequency for each class were calculated, a feature was only considered important for a class, if it's presence in one class is higher than the average relative frequency of that class as well as lower than the average relative frequency of the other class. This work has been reported in our published work
8) Sampling method: A selective oversampling of minority class is introduced in the construction of the models. For each of the prediction end-points, the active (positive) and inactive (negative) data are fragmented using RECAP and ROTBONDS fragmentation methods. The propensity score (PS) for each of the uniquely occurring fragments in both the sets is computed. Only those molecules having the highest propensity scores for fragments conserved for the active class are randomly chosen to be duplicated and added to the original data set. (This in turn reduces the variance). Both steps are repeated until the minority class consists of as many samples as the majority class for all the models.
9) Fragment propensity based CLUSTER cross-validation : Often an entire molecule may not be responsible for the activity, but a local feature (such as a substructure or a fragment) in a molecule may be responsible for the desired response. Chemical fragments are local parts of chemical structures, representing molecular features useful in the modelling of biological or physiochemical properties of chemicals. Therefore, our models take into consideration of local similarity when compared to overall (complete) similarity between set of two active or two inactive compounds for sampling and cross-validation partition. Fragments propensities were mainly used to detect the meaningful features and to capture continuous relationships that exist between the fragments of the same class. They offer intuitive interpretation of the model performance, easy to generate and handle. The 10-fold cross-validation for all the models were performed using fragment- based similarity of compounds. The fragment propensities were calculated for both active and inactive class, as continuous real-valued numbers in the range between 0= low and 1= high. The compounds were thus group into 10 parts based on the fragment propensities. Thus, the group of compounds sharing the fragments propensities were distributed across the folds. The compounds with unassigned fragment propensity were then randomly assigned across the fold. The Compounds assignment to the different folds was done ensuring fragment similarity of compounds and similar ratio of actives to inactives in all the folds, including training and test set.
In our acute toxicity model, we are not only interested in the prediction of one toxicity class, but all of the six toxicity classes. Therefore, we have calculated the sensitivity, specificity and precision for all toxicity classes considered. The overall sensitivity, specificity and precision values have been calculated as averages weighted by the number of compounds in the validation set which belong to a specific class.
For other models, since some of the datasets were imbalanced (having minority and majority class), we have used balanced accuracy and AUC-ROC to access the models)
3.12 Sampling methods
The following are the different data sampling methods as used in this study to handle the imbalanced datasets:1) No Sampling: All the data were used without any manipulation, so called ‘original dataset’.
2) Random Under Sampling (RandUS): The data points from the majority class are removed randomly.
3) Augmented Random Under Sampling (AugRandomUS): Random under sampling in general removes instances of the dataset randomly. In this modified version, the randomness was reduced by utilizing a specifically calculated fingerprint called most common features (MCF) that incorporates all the common features in the data set. The features in this fingerprint are derived from MACCS fingerprints and Morgan fingerprints respectively. To produce this fingerprint the overall average frequency of all the features in the majority class is computed. Then, for each bit position of the fingerprint the relative frequency of ones in the complete data set is computed. If the relative frequency of a bit position is higher than the average frequency the respective bit position and the frequency is saved. Following the average number of features per fingerprint of the majority class is used to specify the number of the features per fingerprint of the MCF fingerprint, whereas the features themselves are specified by the saved features having the highest relative frequencies. Subsequently iteration is performed that is completed as soon as the majority data set is reduced to the size of the minority data set. In each step, a number of samples being the most similar to the MCF fingerprint are collected in a list. Then a number of instances is randomly chosen from the list and removed from the data set. Thereafter, a new MCF fingerprint is computed and the iteration is continued. In this way, the samples most similar to the MCF fingerprint are removed; the loss of variance of the majority set is decreased. In addition, the loss of information is reduced by removing a limited number of samples per calculated MCF fingerprints.
4) Random over sampling (RandOS): Data points from the minority class are randomly chosen and added to the existing minority class.
5) Augmented Random Over Sampling (AugRandOS): Random oversampling in this case also follows the same principle mentioned under the augmented random under sampling before. Only difference in this case, in each iteration step a list of samples most dissimilar to the MCF fingerprint is created. A part of the list is chosen randomly to be duplicated and added to the original data set. Since the samples most dissimilar to MCF are duplicated the loss of variance is relatively low. Both steps are repeated until the minority class consists of as many samples as the majority class.
6) K-Medoids Under Sampling (kMedoids1): K-medoids is a clustering algorithm that is used to under sample the original majority class. A medoid is itself an instance of the majority class utilized as a cluster center that has the minimum average dissimilarity between itself and all majority data points in its cluster. The number of medoids is equal to the number of majority class instances. A sample is assigned to that cluster with which center it shares the highest similarity based on Tanimoto coefficient (Willett, 2003). For each of the medoids the sum of the similarities between itself and all samples belonging to its cluster is calculated. The algorithm tries to maximize the combination of these sums by performing iteration. The iteration is limited to 100 steps, in each of the iterations new medoids are randomly chosen and the overall sum of Tanimoto similarities is calculated. The set of medoids producing the highest sum is used as under sampled majority class. By means of clustering by similarity, this approach creates a subset of which each individual data point represents a group of structurally related molecules, in turn reducing the information lost by under sampling.
7) K-Medoids Under Sampling (kMedoids2): Similarly to kMedoids1 this method starts with randomly choosing n samples as medoids, where n is equal to the number of data points in the minority class. For each of the chosen medoids, a total number of 30 iterations are assigned. In each iterative step, a medoid is exchanged with a random majority class sample, new clusters are computed and the cost is calculated using Tanimoto coefficient. The final set of medoids is chosen based on the maximum sum of similarities.
8) Synthetic Minority Over-Sampling Technique-using Tanimoto Coefficient (SMOTETC): The SMOTE method creates synthetic samples of the minority class to balance the overall data set. Depending on the amount of oversampling a number of samples of the minority class are chosen. For each of those, the k-nearest neighbors are identified, utilizing the Tanimoto coefficient as similarity measure (Willett, 2003). The feature values of the new synthetic data points are set to the value occurring in the majority of the chosen sample and two of its k-nearest neighbors.
9) Synthetic Minority Over-Sampling Technique-using Value Difference Metric (SMOTEVDM): This method is also based on SMOTE, but the k- nearest neighbors are chosen using the Value Difference Metric (VDM) as similarity measure. The VDM defines the distance between analogous feature values over all input feature vectors. More detailed information on the algorithm for computing VDM can be found here (Sugimura et al., 2008).
4. Tutorial
This tutorial shows how to run a toxicity prediction and how to interpret the results. If you have any questions, which are not answered in the FAQs, please feel free to contact us!
Thanks to the YouTube chanel ThePharmaLab, there is also a short video tutorial avaiable.
4.1 How to run a toxicity prediction
To start a toxicity prediction, please go to Tox Prediction. Here, you can either draw your input compound, paste the content of a molfile in textform or search for a compound name online: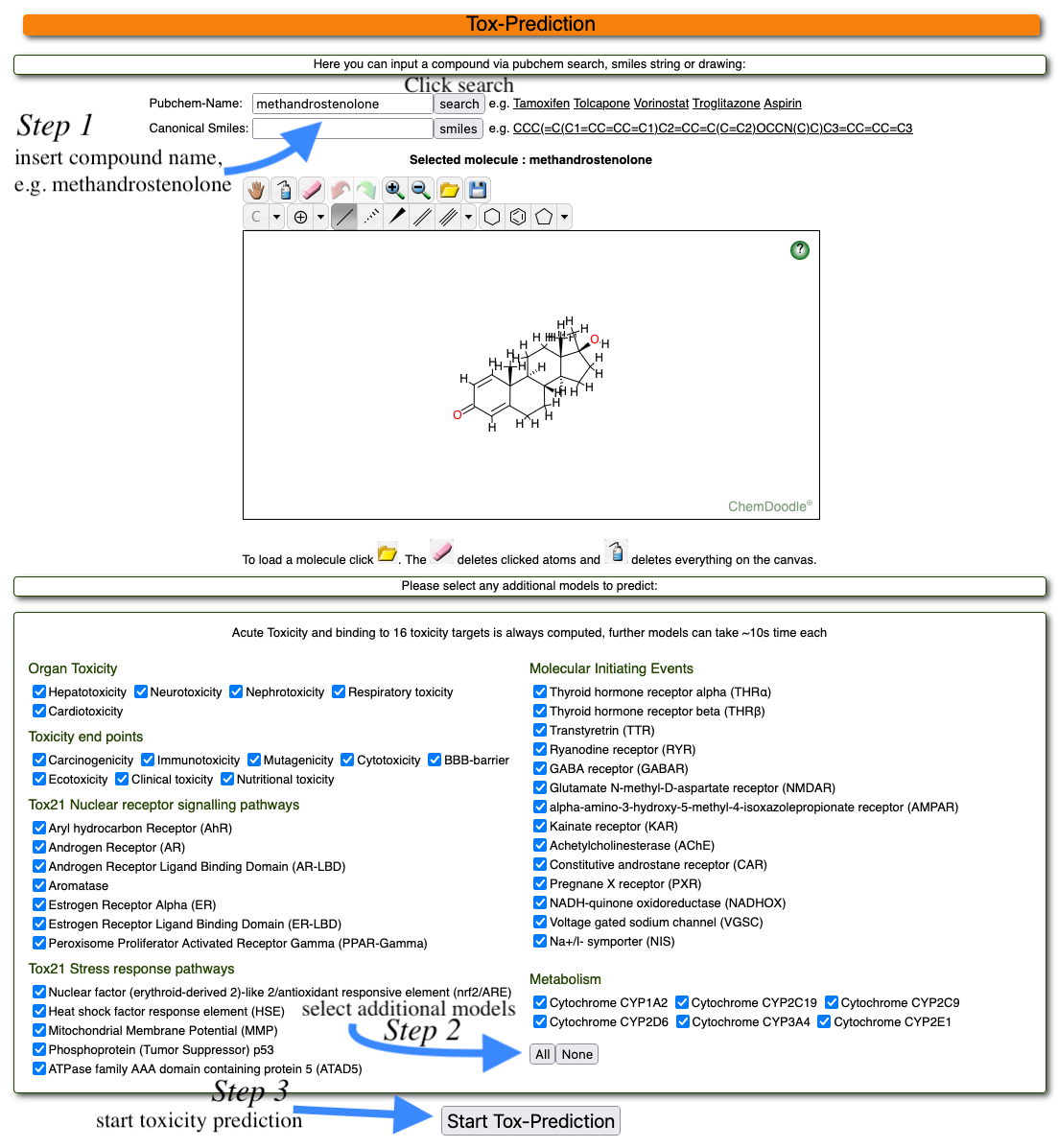
To draw a chemical structure, use the buttons in the second row (as shown above). You can change atom types by clicking on the arrow next to "C" or change bond types or draw ring structures. Please note that a carbon atom is already drawn (grey dot in the middle of the drawing area). To open a molfile, please click on the yellow folder button in the first row (as shown above). You can paste the contents of a molfile (text from) here. You can also search for a known compound online. To do that, click on the binocular in the first row of buttons (as shown above). You can search for a compound name in the PubChem database.
To illustrate the functionalities and possible application of the ProTox 3.0 web server, methandrostenolone was selected. Methandrostenolone is an orally active anabolic androgenic steroid. It was introduced to the market in 1960. Later, was withdrawn in the year 1982 from different countries – France, Germany, UK, USA; reasons for withdrawal were cited as off-label abuse.
To use the example compound, simply type the name and click on name search Start Tox-Prediction. To clear the drawing area, press the button with the blue bottle in the first row of buttons.
Once you have drawn or inserted an input molecule, you can start the toxicity prediction by clicking on the Start Tox-Prediction button below the drawing area. Additionally, you can select the models of your choice or all the models mentioned for prediction. If you are only interested in acute toxicity and toxicity targets of your chemical compounds, the server by default calculates that.
Please note that the prediction of toxicities of multiple compounds can be time-consuming. An estimation of the calculation time is given at the top of the results page, but in case the user does not want to wait for the results, the results page can be bookmarked and the results accessed at a later time. The results (predicted LD50 values, predicted toxicity class, etc.) are given in a tabular format. Further details, including similar compounds with known toxic class and possible toxicity targets, Toxicity model report can be obtained for each compound by clicking on the 'plus button'.
Using our ProTox 3.0 prediction pipeline, Methandrostenolone has been predicted with Toxicity class 4 for acute oral toxicity with LD 50 value of 1000mg/kg, with a prediction accuracy of 100.00%. Three structurally similar compounds and their physicochemical properties distribution plots are reported from the database. The drug was predicted to be active for liver toxicity, neurotoxicity, and respiratory toxicity. It was also predicted to be active for the BBB permeability, immunotoxic, and clinically toxic under the toxicological endpoints class. Four toxicological pathways – NR-AR, NR-AR-LBD, NR-ER, and NR-ER-LBD were predicted active with a high probability of 1.0. One MIEs endpoint -Ache was predicted as active for this drug. The metabolic enzyme CYP2C9 was predicted to be active for the drug.
Additionally, four different toxicity targets (Androgen Receptor, Amine Oxidase A, Glucocorticoid Receptor, and Progesterone Receptor A) are predicted with probable binding.
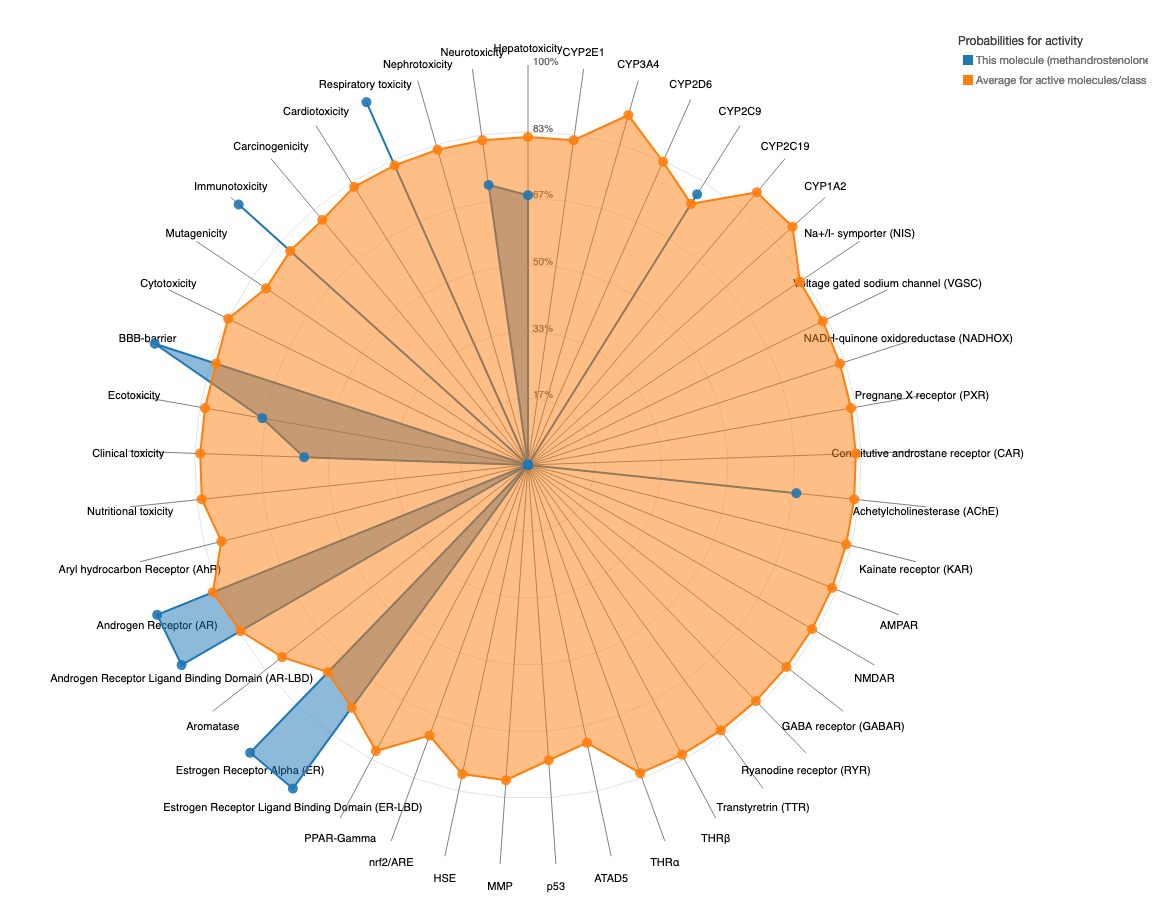
A toxicity radar plot (example below) is provided to assess the comparison between the different toxicity models active compounds average probability from the training set to that of the input compound. The plot can be accessed using the 'Open Toxicity Radar Chart' link that will appear on top of the page once the computation is complete, which will open the chart in a new tab. The toxicity profile of the input compound is shown using blue lines/dots which represents the predicted probabilities of the input compound for respective ProTox 3.0 models. The data displayed is orange dots/lines is the average probability of its active class, acquired by computing from the training set data for each model (see model info). For the example case Methandrostenolone, the predicted probabilities for the models respiratory toxicity, immunotoxicity, BBB-barrier, androgen receptor, AR-LBD, CYP2C9, estrogen receptor alpha and ER-LBD is higher than the average probabilities for each of these models, representing a strong prediction confidence. However, the prediction probability for e.g. the Ecotoxicity model is lower than the average probability of the training set active compounds. This chart helps the user to get an understanding, how strong is the overall prediction of the input compound, considering its activity for multiple toxicity endpoints.
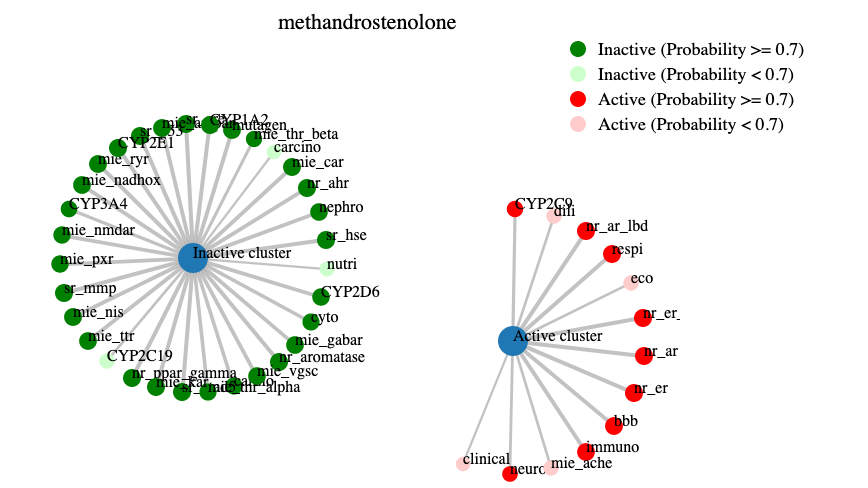
Additionally a network plot (example below) was drawn to give a fast overview over the active and inactive cluster.
4.2 Toxicity prediction results
The results of the toxicity prediction will open automatically after a few moments. The report for your input compound will look like this: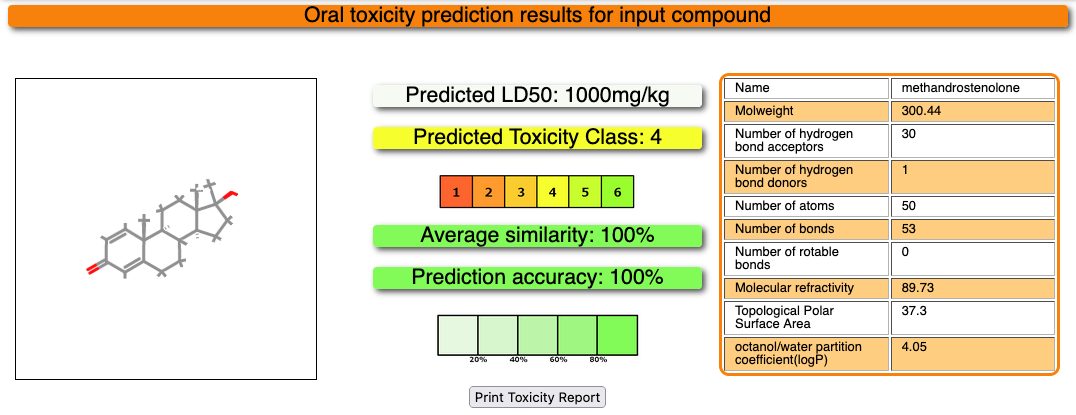
The structure of your input compound is shown in the box on the left whereas some properties of the input compound are displayed in the table on the right. The prediction results are shown in the middle of the page. A prediction for the median lethal dose (LD50) is given in mg/kg body weight at the top, followed by the toxicity class. 6 different toxicity classes are distinguished, as explained here, and each class is displayed in a different color (see box).
Furthermore, a prediction accuracy is calculated and displayed. The more saturated the prediction accuracy box color, the higher the accuracy (see box below). The prediction accuracy depends on the similarity of the input compound to compounds with known LD50 values as well as the hit rates obtained in a cross-validation study.
A model report can be easily be downloaded and printed using the Print Toxicity Report button.
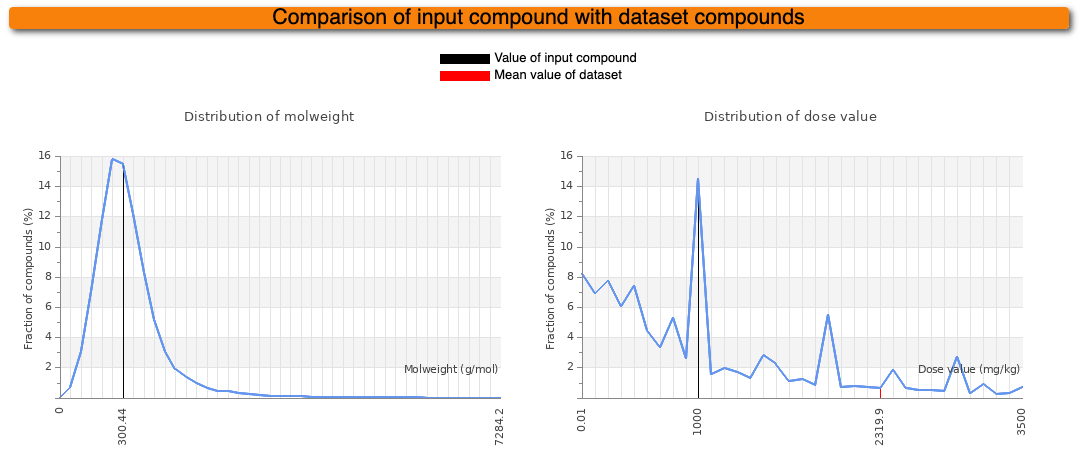
In addition to the prediction results, some information about the input compound is given. The diagram on the left indicates the molecular weight (MW) distribution of compounds in our dataset. The mean MW is indicated as red line whereas the MW of the input compound is indicated as black line. In the diagram on the right, the distribution of LD50 values of our dataset is shown. Again, the mean of our dataset is shown in red and the predicted median lethal dose of the input compound is shown in black.
Below the diagrams, 3 compounds which are the most similar compounds of our dataset to the input molecule are displayed. Their chemical structures as well as their properties are shown. Please note that the toxicity class is assigned based on 3 different schemes. First of all, if multiple LD50 values are available for one compound, the toxicity class can be assigned based on the minimum (min) or the average (avg) dose. Secondly, the toxicity class can be calculated based on the concentration, taking into account the molecular weight of the compound.
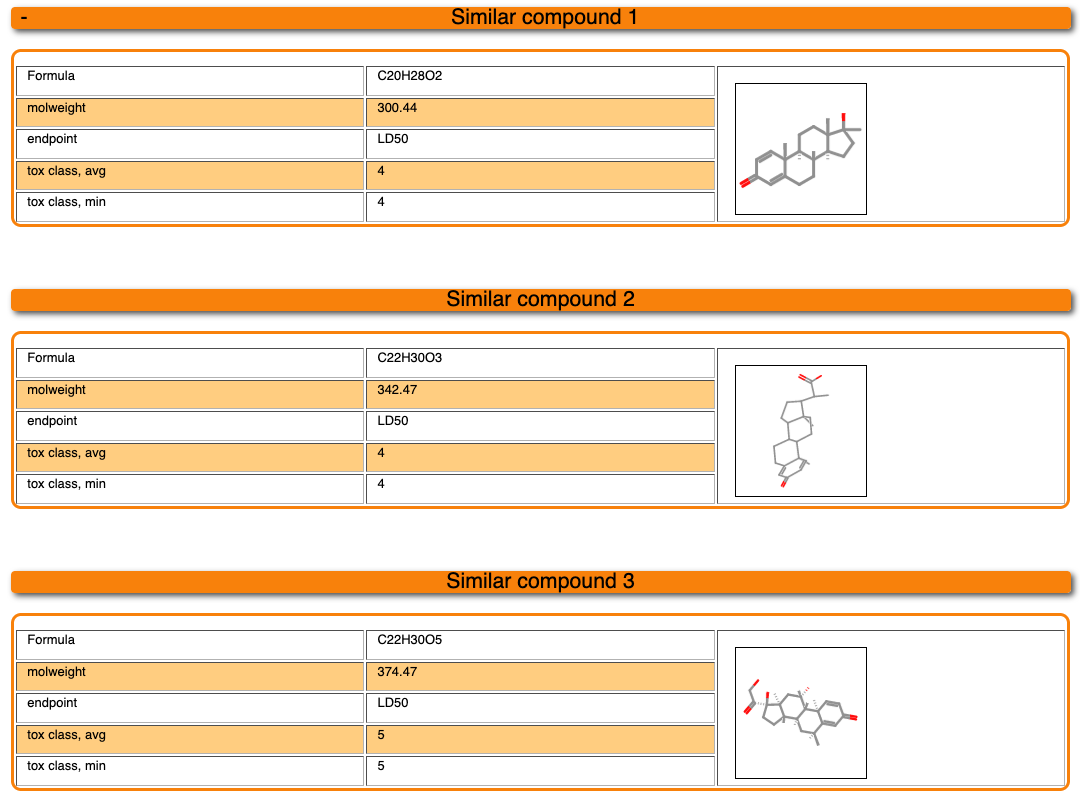
4.3 Toxicity model report
The toxicity model report gives an overview of the predicted activity computed by the various machine learning models. After the calculations are completed for multiple models, the result looks like below :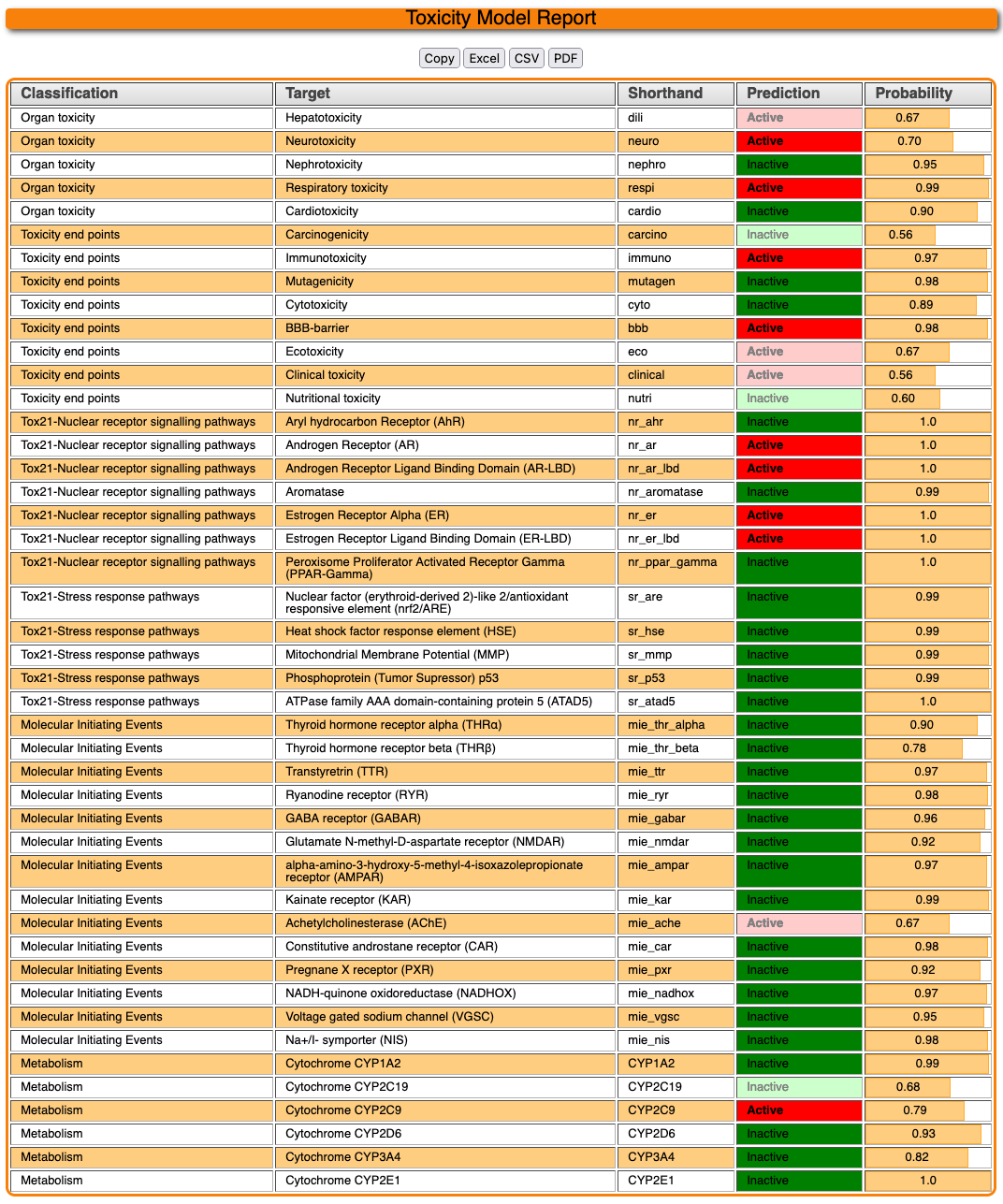

Targets are sorted by the leftmost classification. A target predicted to be active with the input molecule will be emphasized by a bolded prediction tag. The probability on the left hand side gives a confidence estimate for the prediction. Data points with a confidence below 70% (0.7) are normally omitted, displaying a value of "Below Threshold". While the model is still computing, a value of "Calculating..." is shown here. The top of the page should give an estimated of when the models are finished, but at most, it should take around 4 minutes, less if part or all of the selected models already has been precomputed in the past.
The result table can be copied or downloaded in various formats like excel file or CSV using the buttons above.
4.4 Toxicity target indication
The results of the toxicity target alert will show up at the toxicity prediction report page and will look similar to the image below:
The first table gives an overview over all investigated toxicity targets. The target name abbreviations are given in the first row and they contain hyperlinks to further information about the protein targets. The colors indicate how probable binding to the toxicity target is: black indicates no binding, whereas yellow, orange and red indicate possible binding. The more intense the color, the more probable the binding is.
If toxicity targets are found for an input ligand, a second table provides the details of the targets found. The target name as well as its average pharmacophore fit and average similarity to known ligands of this target (based on Tanimoto similarity) are given. The average pharmacophore fit indicates how well compounds similar to the input compound can fit the protein-ligand-based pharmacophores developed for every target. The average similarity, on the other hand, indicates the similarity of the input compounds to molecules which have been shown to bind at this particular target.
Last updated: May 2024
Disclaimer: Compound structures submitted will not be released under any circumstances. This work is licensed under a Creative Commons License.
Disclaimer: Compound structures submitted will not be released under any circumstances. This work is licensed under a Creative Commons License.